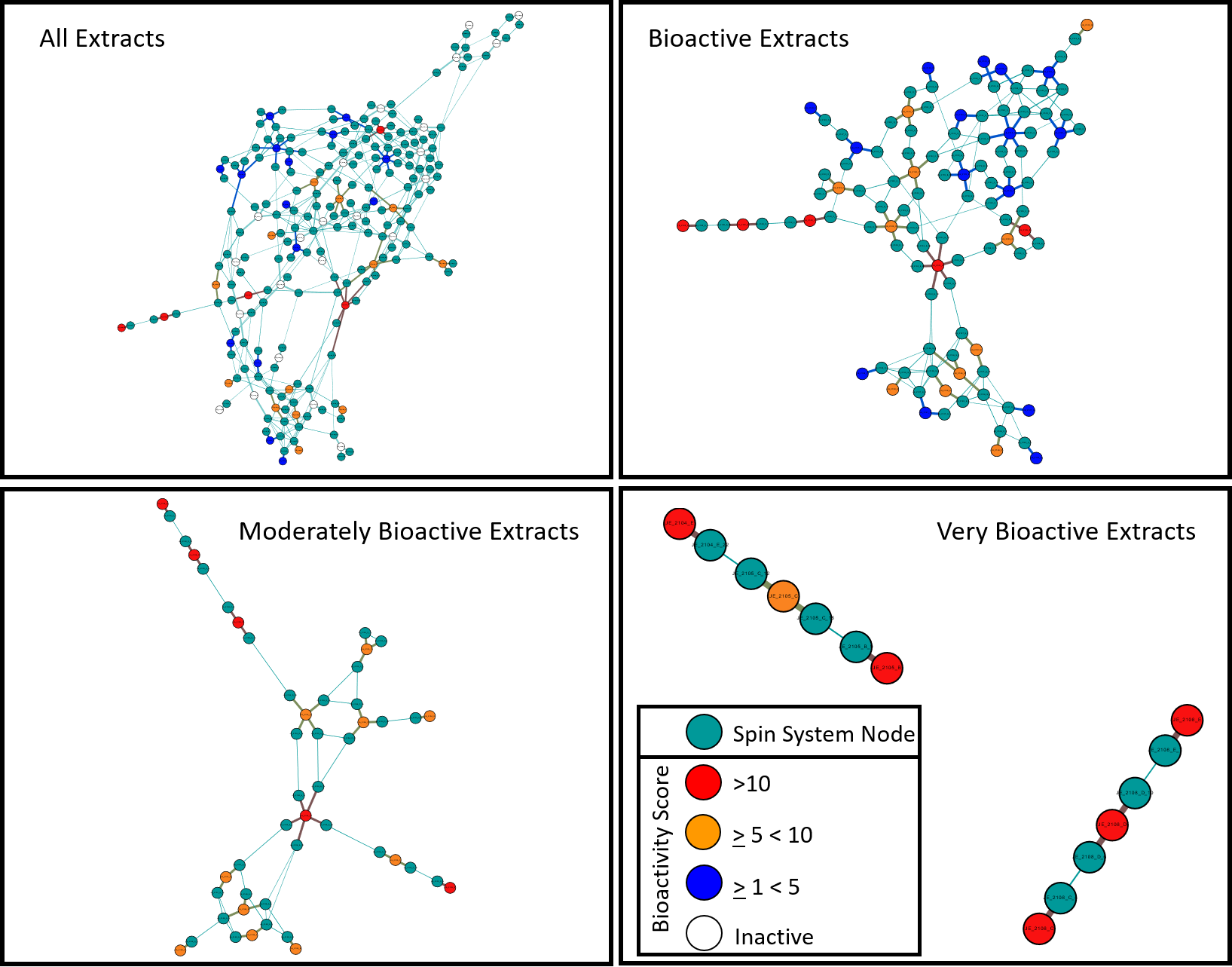
Bioactivity Integration
MADByTE can inform an investigator about either shared structural elements or novel structural motifs that can be found across a large dataset. In untargeted metabolomics, using these shared structural motifs allows investigators to create informed hypotheses about which chemical entity may be of importance. Once networks are constructed, layering other sample relevant data, such as bioactivity, allows for prioritization of bioactive metabolites that are observable as features. All extracts that were analyzed via MADByTE that were also screened in the BioMAP (doi: 10.1016/j.chembiol.2012.09.014) platform were filtered, processed, and color coded via the bioactivity ‘hit’ rate.
In the case of one of these networks, these findings allowed for the isolation of the bioactive constituent from these extracts to be isolated, and submitted for re-screening, was proven to be both the driving force of the bioactivity profile of the extract, as well as the driving force of the structural relatedness network clustering.
Vendor independent integration
Over the last few months, we have been working on the adaptability of MADByTE to make it easy for others to use, regardless of workflow. Currently, the system is set up to parse through and interrogate Bruker data automatically. However, if you’re like most end users, you may be more comfortable with MestRe Nova, ACD, or Delta. To make it easy, we’ve been working behind the scenes in getting MADByTE to fetch these data for you so you don’t have to convert things around.